|
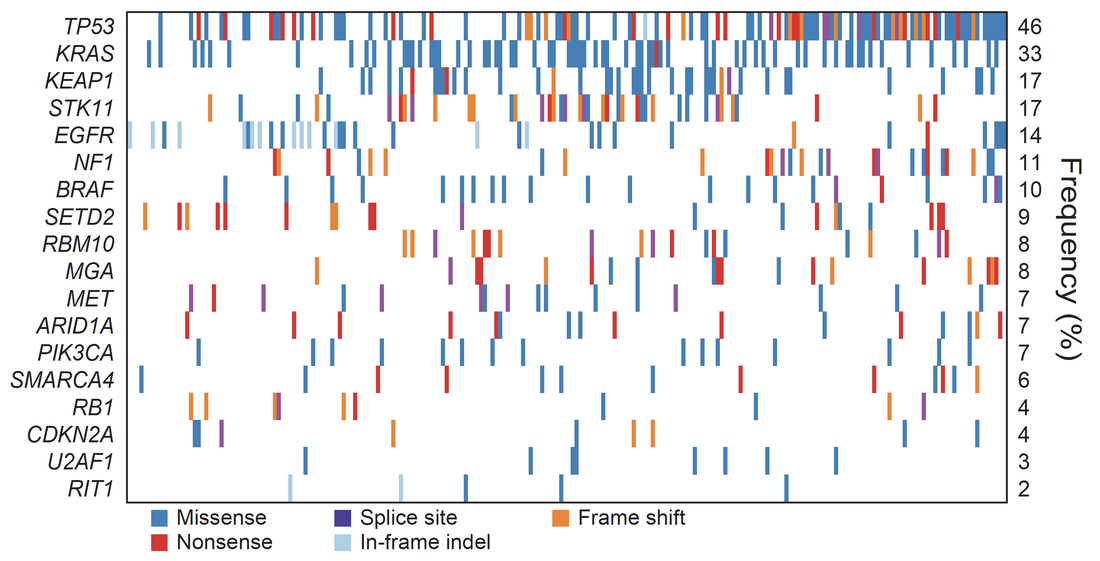
Co-mutation plot from whole exome sequencing of 230 lung adenocarcinomas.
TGCA, Nature, 2014. Comprehensive molecular profiling of lung adenocarcinoma. |
Genomics of Lung and Other Cancers
Our work focuses on discovery of cancer driver genomes, structural analysis of cancer genomes, and the link between germline and somatic mutations in cancer.
Discoveries of cancer driver genes
The recognition that kinase inhibitors can be critical for treating human cancer focused our initial lung cancer research efforts on identifying mutations in protein kinase genes, beginning with the discovery of somatic BRAF mutations in lung adenocarcinoma (Naoki et al., 2002). Working with colleagues at DFCI, we were the first to identify activating mutations in EGFR (Paez, et al., 2004) in lung cancer and to correlate these alterations with patient response to the kinase targeting drug, gefitinib. In the years since, we have discovered numerous such recurrent alterations in kinase genes in lung cancer, e.g. FGFR2 and FGFR3 (Liao, et al., 2013) and DDR2 in squamous cell lung carcinoma (Hammerman, et al, 2011), and RAF1 (Imielinski, et al., 2014) in lung adenocarcinoma.
We have identified multiple genes subject to recurrent driver mutations in lung adenocarcinoma, including the U2AF1 splicing factor gene and the putative splicing factor, BRM10 in lung adenocarcinoma (Imielinski, et al., 2012). Through our studies of lung cancer genomes as part of The Cancer Genome Atlas, we discovered loss of function mutations in the HLA-A gene in squamous cell lung carcinoma (TCGA, 2012) and in the MGA tumor suppressor gene in the MYC pathway in lung adenocarcinoma (TCGA, 2014), as well as PPP3CA, DOT1L, and FTSJD1/CMTR2 in lung adenocarcinomas, RASA1 in squamous cell carcinomas, and KLF5, EP300, and CREBBP in both lung cancer subtypes (Campbell, et al., 2016).
Genome structural alterations
Our work on structural alterations has evolved from SNP array studies of copy number, identifying focal amplifications of TERT, NKX2-1 (Weir et al., 2007), and SOX2 (Bass et al., 2009) in lung cancers, to a new focus on whole genome sequencing and now long-read sequencing.
We discovered super-enhancer amplifications targeting MYC in lung adenocarcinoma and other cancer types (Zhang, et al., 2016), KLF5 in lung squamous cancer and other cancer types (Zhang, et al., 2018), and androgen receptor in prostate cancer (Viswanathan et al., 2018). We also discovered somatic hotspot indels in non-coding regions of lineage-defining genes in cancer, such as surfactant protein genes in lung cancers (Imielinski, et al., 2017).
Our current focus is on applying and improving long read sequencing for analysis of cancer somatic structural variants, beginning at the telomere (Tan et al., 2022).
Germline risk for somatic mutations in cancer
When we discovered somatic EGFR mutations in lung adenocarcinoma (Paez et al., 2004), we were surprised to find that the mutation frequency is ~50% in cancers from East Asian patients and ~10% in cancers from patients of European origin. Further studies revealed that patients of African descent also have about a ~10% somatic mutation frequency in EGFR (Campbell et al., 2017).
Recently, we have studied lung cancer somatic mutations in patients from Mexico and Colombia that suggest a germline link to both EGFR and KRAS mutation frequencies, where the fraction of Native American ancestry correlates with EGFR mutations and anti-correlates with KRAS mutation (Carrot-Zhang et al., 2021). We are looking for the germline locus or loci underlying this association—nothing yet but we are trying hard!
Discoveries of cancer driver genes
The recognition that kinase inhibitors can be critical for treating human cancer focused our initial lung cancer research efforts on identifying mutations in protein kinase genes, beginning with the discovery of somatic BRAF mutations in lung adenocarcinoma (Naoki et al., 2002). Working with colleagues at DFCI, we were the first to identify activating mutations in EGFR (Paez, et al., 2004) in lung cancer and to correlate these alterations with patient response to the kinase targeting drug, gefitinib. In the years since, we have discovered numerous such recurrent alterations in kinase genes in lung cancer, e.g. FGFR2 and FGFR3 (Liao, et al., 2013) and DDR2 in squamous cell lung carcinoma (Hammerman, et al, 2011), and RAF1 (Imielinski, et al., 2014) in lung adenocarcinoma.
We have identified multiple genes subject to recurrent driver mutations in lung adenocarcinoma, including the U2AF1 splicing factor gene and the putative splicing factor, BRM10 in lung adenocarcinoma (Imielinski, et al., 2012). Through our studies of lung cancer genomes as part of The Cancer Genome Atlas, we discovered loss of function mutations in the HLA-A gene in squamous cell lung carcinoma (TCGA, 2012) and in the MGA tumor suppressor gene in the MYC pathway in lung adenocarcinoma (TCGA, 2014), as well as PPP3CA, DOT1L, and FTSJD1/CMTR2 in lung adenocarcinomas, RASA1 in squamous cell carcinomas, and KLF5, EP300, and CREBBP in both lung cancer subtypes (Campbell, et al., 2016).
Genome structural alterations
Our work on structural alterations has evolved from SNP array studies of copy number, identifying focal amplifications of TERT, NKX2-1 (Weir et al., 2007), and SOX2 (Bass et al., 2009) in lung cancers, to a new focus on whole genome sequencing and now long-read sequencing.
We discovered super-enhancer amplifications targeting MYC in lung adenocarcinoma and other cancer types (Zhang, et al., 2016), KLF5 in lung squamous cancer and other cancer types (Zhang, et al., 2018), and androgen receptor in prostate cancer (Viswanathan et al., 2018). We also discovered somatic hotspot indels in non-coding regions of lineage-defining genes in cancer, such as surfactant protein genes in lung cancers (Imielinski, et al., 2017).
Our current focus is on applying and improving long read sequencing for analysis of cancer somatic structural variants, beginning at the telomere (Tan et al., 2022).
Germline risk for somatic mutations in cancer
When we discovered somatic EGFR mutations in lung adenocarcinoma (Paez et al., 2004), we were surprised to find that the mutation frequency is ~50% in cancers from East Asian patients and ~10% in cancers from patients of European origin. Further studies revealed that patients of African descent also have about a ~10% somatic mutation frequency in EGFR (Campbell et al., 2017).
Recently, we have studied lung cancer somatic mutations in patients from Mexico and Colombia that suggest a germline link to both EGFR and KRAS mutation frequencies, where the fraction of Native American ancestry correlates with EGFR mutations and anti-correlates with KRAS mutation (Carrot-Zhang et al., 2021). We are looking for the germline locus or loci underlying this association—nothing yet but we are trying hard!